Simulate gene data of PSD model including P, F, G.
Usage
psd_simulation(
I,
J,
K,
type.id = c("A", "B"),
parm_alpha = 0.1,
parm_sd = 2,
parm_F = NULL,
data = data_HGDP
)Arguments
- I
The number of individuals to simulate.
- J
The number of SNPs to simulate.
- K
The number of populations to simulate.
- type.id
Choose type. Should be one of
"A","B".- parm_alpha
A parameter of the normal distribution in the simulation of the first type of P.
- parm_sd
A parameter of the normal distribution in the simulation of the second type of P.
- parm_F
A parameter of the beta distribution in the simulation of F. It has the same length as
K.- data
The data needed to be provided in order to collect the suballele frequencies of real SNPs during the simulation of F.
Value
A List with the following parameters:
GThe I x J simulated matrix of counts.
PThe I x K simulated population scale matrix of the individuals.
FThe K x J simulated gene scale matrix of the populations.
Examples
data_simuA <- psd_simulation(60, 250, 3, type.id = "A", parm_F = c(0.1, 0.05, 0.01))
plot_structure(data_simuA$P, pops = rep(3:1))
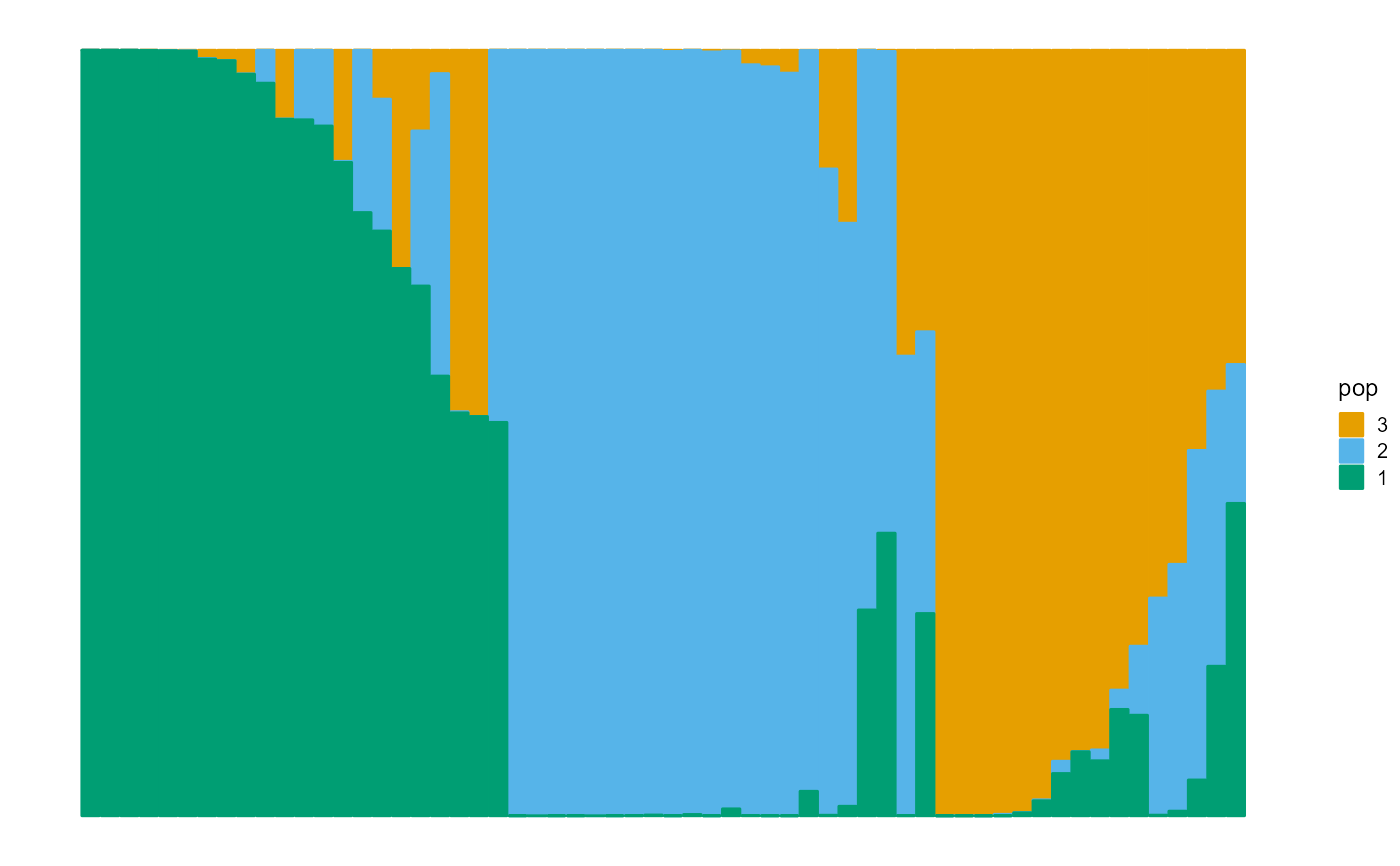 data_simuB <- psd_simulation(100, 500, 5, type.id = "B")
plot_structure(data_simuB$P, pops = rep(5:1))
data_simuB <- psd_simulation(100, 500, 5, type.id = "B")
plot_structure(data_simuB$P, pops = rep(5:1))

